CIMI Modeling Architecture Guide |
Version 0.09 |
Published Date: 03/26/2017 Authors: HL7 CIMI Work Group |
The CIMI Model’s Alignment to Terminology 12
A High Level View of the CIMI Logical Model 12
What is a ‘reference model pattern’ or ‘clinical pattern’ 14
The CIMI Archetype Hierarchies 14
The CIMI Core Reference Model Module 16
The LOCATABLE and ASSOCIATION_CLASS Root Types 19
The CIMI Foundational Reference Model Module 20
The CLUSTER/VIRTUAL_CLUSTER hierarchies 22
The PARTY, PARTICIPATION, and PARTY_RELATIONSHIP pattern 22
The CIMI Clinical Reference Model Module 23
The CIMI Party and Participation Patterns 23
The CIMI Clinical Statement Pattern 24
The CIMI Attribution/Provenance patterns 30
The CIMI Assertion and EvaluationResult Pattern 32
Common CIMI Data Structures 38
An Introduction to CIMI Archetype Hierarchies 42
The Attribution archetype hierarchy 43
The StatementContext hierarchy 44
The StatementTopic Hierarchies 45
The Clinical Statement Hierarchies 47
A Note About Other Cluster Archetype Hierarchies 51
Top Level Value Set Constraints 51
Archetypes constraining attribute types 52
Version | Date | Author | Amendment History |
0.1 | 2012-09-13 | Linda Bird | First draft for comment |
0.2 | 2012-10-26 | Rahil Qamar Siddiqui | Revised section 6.6 on Terminology Binding |
Added sub-sections 6.6.1 to 6.6.4 with preliminary details on the main types of terminology bindings within scope for CIMI. More details to be added in due course. | |||
.03 | 2016-11-16 | Susan Matney | Draft for ballot comment |
.04 | 2016-11-23 | Claude Nanjo | Integration of architectural sections |
05 | 2016-11-25 | Susan Matney | Edits |
0.6 | 2016-11-27 | Jay Lyle | Edits |
0.7 | 2016-11-29 | Claude Nanjo | Review of style guide section and edits |
0.8 | 2017-03-15 | Claude Nanjo | Review of comments and final updates to the documentation based on new artifacts. |
0.9 | 2017-03-22 | Claude Nanjo | Incorporating Patrick’s changes |
Figure 1: Version History
The Clinical Information Modelling Initiative (CIMI) is a Health Level Seven (HL7) workgroup dedicated to providing a common definition of health information content so semantically interoperable and computable information may be created and shared in health records, messages, and documents.
In achieving this goal, CIMI has established a CIMI modelling methodology, style guide, and a set of models which together demonstrate and test the approach to CIMI clinical modelling.
CIMI modelling is the overall strategy for specifying, at a granular level and in a computable fashion, the structure and semantics of the common data elements that will be stored in an Electronic Health Record (EHR). More exactly, the CIMI Model specifies how to create a “model” of a particular type of element (e.g., a heart rate measurement, a laboratory observation, or a procedure performed). The model declares how a valid “instance” of the element should be structured, and the semantics of the structure. By analogy, a blueprint is used for building cars. All the cars built as a result of the blueprint conform to the specifications of the blueprint. The cars conforming to the blueprints are analogous to instances of data (e.g., John Smith’s heart rate measurement at 11 am on January 26th, the observation of Mary Black’s blood glucose on March 20th, or the procedure performed on Jeff Brown June 23rd at 10 am), while the blueprints are analogous to the models (e.g., a model for heart rate measurement instances, a model for laboratory observation instances, or a model for procedure performed instances).
The purpose of this document is to give modellers practical information needed to author CIMI models. This document describes some initial proposals by the CIMI modelling task force to help inform CIMI’s model architecture, modelling methodology, and style guide.
The CIMI Logical Model and the FHIR Resource Model are complementary standards. Unlike FHIR, CIMI does not provide a specification for the representation of instances of clinical data. CIMI provides source-content for the production of implementation models such as FHIR. The CIMI Model offers a formal specification architecture for a set of consistent FHIR logical, resource, and extension profiles to enable both the interoperable exchange of clinical information and the queries, analyses, and computations performed upon such information. Services whose APIs make use of these FHIR profiles will be able to leverage the expressivity and computability of the CIMI logical model within health applications.
The CIMI logical model offers a stable, consistent, and computable clinical model for the development of knowledge artefacts and for the design of execution systems making use of these models in clinical logic. Clinical Decision Support and Clinical Quality Measures are particularly dependant on the computability characteristics targeted by CIMI.
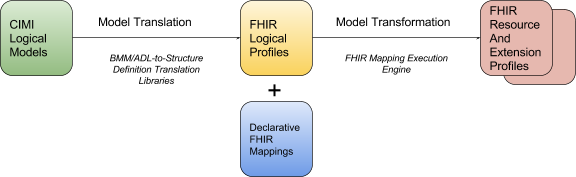
In order to achieve this, the CIMI preferred models must first be transformed into FHIR resource profiles. The CIMI logical model provides the underlying specification for such profiles through the translation of both reference model structures and archetype constraints into their equivalent representations in corresponding FHIR structure definitions. Once the set of CIMI-FHIR profiles has been generated, FHIR instances, conformant with these CIMI profiles, represent conformant CIMI instances.
Figure 2: FHIR Profile Generation from CIMI Models
Given their differing requirements, CIMI models and FHIR resources may differ in their structure and therefore transformation costs are inevitable. For instance, while CIMI clinical statements are compositional structures to support model reuse and consistency, FHIR resources tend to prioritize ease of implementation, and tend to be flatter. In order to minimize these transformation costs, CIMI aims to:
At a high level, the transformation of CIMI models into FHIR profiles can be achieved as follows:
Archetypes in CIMI are defined hierarchically with more specific archetypes further tightening the constraints of their ancestors in the hierarchy. In order to retain this structure in FHIR, the translation of CIMI archetype hierarchies into FHIR resource profiles will most likely result in the generation of layered resource profiles in FHIR that mimic the structure found in CIMI.
One important aspect to note is FHIR’s architecture and the semantics of FHIR Profiles are tightly focused on the needs of an information model transport layer. FHIR’s success depends on the successful implementation of FHIR across a broad and diverse ecosystem of clients and servers. In the FHIR community, the weighing of tradeoffs between sophistication and simplicity trend heavily towards simplicity. FHIR depends on the authoring process to deliver consistent and computable clinical information. How this is achieved is out of scope. In contrast, the goal and scope of CIMI is tightly focused on providing standards, patterns, and tooling to deliver this process.
CIMI models can be applied beyond FHIR implementations, but given the immediate relevance of FHIR within the clinical community, the production of FHIR Profiles from CIMI clinical models is being given the highest possible priority. The CIMI workgroup believes the value of FHIR resources and profiles generated from CIMI clinical models offers the best of both worlds: the clarity, consistency, and computability of CIMI and the implementation simplicity of FHIR.
In addition to existing collaborations with the FHIR, CDS, CQI and Patient Care Working Groups, the CIMI team intends to approach other working groups at HL7 in order to support CIMI harmonization with existing standards and encourage stronger working group coordination.
CIMI and V3 share the goal of creating models that support interoperability. CIMI has profited from many of the core concepts in the V3 RIM, including the V3 data types and foundational classes for entities, acts, and participation. The V3 process used HL7 developed tools and processes to specialize the RIM classes to RMIMs, and then further specialize the RMIMs to create implementable message definitions. CIMI starts with a core reference model that is entirely general, and then builds high level patterns for common classes of healthcare data. These patterns are at roughly the same level as FHIR resources. CIMI then uses formal health information modeling languages (Archetype Definition Language and Archetype Modeling Language) to constrains the patterns to definitions of very specific and explicit health care data elements (systolic blood pressure, heart rate, serum sodium level, pain severity, etc.), primarily by specifying standard codes from SNOMED CT, LOINC, and RxNorm. Thus, though there are many common ideas and themes between CIMI and V3, there is very little similarity in the technology, processes, or final content.
The motivation for this informative ballot submission is to solicit further feedback on the core architecture of the CIMI logical model. Given the scope of this effort, the CIMI Team has focused primarily on the following areas:
While this submission describes additional models, including a number of supporting structures, many of these structures have not received adequate internal review at this time and should still be treated as work-in-progress. Therefore, we encourage the reader to prioritize review on the items listed above, though we certainly welcome feedback on any part of the model.
It is also important to note while some value set bindings will be provided as part of this submission, much work still remains to be done. Terminology alignment will be addressed in future ballot submissions and all terminology bindings, apart from those specifying the alignment of model attributes to SNOMED CT Concept Model attributes, are provided as examples at this time.
While great effort has gone into this phase of the CIMI model development and in the preparation of the May 2017 ballot material, the CIMI Working Group recognizes much work still remains. In the months following the May HL7 Working Group Meeting, the CIMI working group intends to address community comments and refactor the model accordingly. By the September 2017 HL7 Working Group Meeting, our goal is to complete the CIMI Reference Model, CIMI top-level archetypes including archetypes capturing US Core and QI Core requirements, and ballot the model as an Informative Specification. By the January 2017 HL7 Working Group Meeting, we intend to complete the terminology bindings for all proposed archetypes, implement detailed clinical models for selected use cases, and specify formal declarative mappings from CIMI to FHIR using the FHIR Mapping Language. The model will then be balloted as a Standard for Trial Use.
We encourage the community to comment on any aspect of the proposed model. In particular, we would like to solicit comments and feedback in the following areas:
CIMI Clinical Models are defined through two closely integrated OpenEHR syntaxes:
BMM is used to define the structural content of components (i.e., the classes and attributes that make up the model), and has sophisticated capabilities to leverage inheritance, composition, and generics, which offers authors the maximum ability for reuse and consistency across definitions. BMM is used to produce three structural layers of schematic definitions.
The three layers of BMM schema consist of:
BMM and it’s three layers of definitions delivers a rich hierarchy of information metadata to allow developers of clinical authoring tools and run-time execution engines opportunity to leverage code-generation to dynamic run-time techniques to deliver sophisticated tools with a high degree of insulation from evolutionary changes in the information schema. BMM is not a technology typically exposed to a Subject Matter Expert (SME) as they author clinical content. It is simply a serialization standard used for standardized persistence and exchange of clinical patterns.
Archetype Definition Language (ADL)
CIMI believes a finite and manageable set of reference clinical patterns can support a much larger number of specific clinical situations recorded in the medical record. It would be daunting to implementors if each bone in the human body had it’s own information model definition. Instead, detailed models for broad clinical domains can be derived from a much smaller set of underlying BMM structural pattern definitions through the application of terminology constraints. For instance, a very large number of physical evaluation results can be derived from only a few reference clinical patterns. The models used to represent such clinical domain scenarios are referred to as Detailed Clinical Models (DCMs) and are defined using the Archetype Definition Language (ADL).
Figure 3: Creation of Detailed Clinical Models (DCMs) from CIMI Reference Model Patterns
When the logical-models are transformed into a specific implementation, implementers get to choose the level of granularity for their information model layer. Using the Fracture topic as an example, the implementation team may choose to store all fractures in a single relational table, even though there may be hundreds of Archetypes defined for the hundreds of bones within the human body. The hierarchy of Archetypes would define all the complex variations of recordable qualifiers and modifiers applying to the different bones. But in this example, each of these variations would be stored in the same schema structure within the information-model.
Archetypes provide the constraints determining what qualifiers and modifiers are valid for very narrow clinical situations. Pelvic fractures and Spine fractures are recorded using a different collection of qualifying elements - yet each is a fracture. ADL provides an expressive collection of semantic options to define the specific clinical situations. But similarly to BMM, ADL is intended to be a serialization format used by the tooling layer to exchange standardized clinical artifact constraints. It would not typically be exposed to SMEs during the authoring process, only to the engineers involved in developing authoring and run-time tools and engines.
BMM / ADL relationship to previous and existing modeling standards:
The most commonly understood and used general-purpose modeling language today is Universal Modeling Language (UML). Two other modeling formalisms used similarly within the Healthcare space are ISO 13606 and openEHR. Several proprietary clinical modeling languages have also been developed internally within individual healthcare systems such as the Clinical Element Modeling language (CEML) from Intermountain. Each of these has its strengths and limitations and none of them are likely to disappear from the landscape in the near future. Therefore, a great deal of effort has been invested in ensuring BMM and ADL formats are semantically compatible with these other formats and in transformation tooling to supporting an ecosystem of intercompatibility between syntaxes.
More will be written about this over time, but the fundamental goal of CIMI is to deliver interoperability between teams who are deeply invested in UML as well as teams leveraging BMM / ADL directly.
CIMI Detailed Clinical Models (also referred to as archetypes) are defined as a set of constraints on the CIMI reference model structural patterns. This reference model is the ‘common language’ used to describe all clinical models. CIMI Clinical models are defined using a constraint language whose semantics are defined by the Archetype Object Model (v2)[1]. These constraint semantics can be represented using either the Archetype Definition Language (ADL)[2] (a direct serialisation of the AOM) or Archetype Modelling Language[3] (AML) (a profile of the Unified Modelling Language). CIMI Clinical models will usually include both semantic bindings (i.e. bindings to terminology expressions that define the meaning of the relevant parts of the model) and value bindings (i.e. bindings to terminology reference sets that define the valid values that may populate appropriate parts of the model). For this ballot cycle, we are interested in semantic bindings to the SNOMED CT Concept model--for the value bindings, we welcome but do not solicit comments.
It is anticipated CIMI Clinical models will be both specialised and extended to develop realm-specific clinical models which will then be further transformed into implementation-specific artifacts.
The following principles guide CIMI’s modelling approach:
Information models are often developed independently of clinical ontologies. As a result, many information models align poorly with the terminologies or ontologies upon which they ultimately depend for their formal semantics. Moreover, by not explicitly specifying the model’s semantics the meaning of the model is left open for interpretation during implementation further hindering interoperability. In an effort to better align models of use with models of meaning, the Clinical Information Model is designed to align closely with the SNOMED CT Concept Model wherever such opportunities exists. In CIMI, the model’s formal semantics are specified through terminology bindings defined at the archetype level. These terminology bindings occur at three levels:
| Anatomical or acquired body structure | 442083009 (<<)
The CIMI model consists of structural patterns and the constraints applied on those patterns:
The CIMI model is persisted in a format conforming to two OpenEHR specifications:
The CIMI model follows a strict rule in the usage of the above-mentioned two OpenEHR specifications in order to cleanly delineate the boundary between the reference model and the archetype hierarchies. The basic meta-modeling language is used to specify the classes, attributes, and relationships making up the model. The archetype definition language is used to define the constraints on the reference model but shall not be used to define new model classes and attributes. In other words, the CIMI model specifies classes and attributes explicitly in the reference model and does not offer a way to extend the model within archetypes.
This approach represents a departure from the approach taken by OpenEHR, which allows the definition of classes and attributes within archetypes using the meta model constructs of ITEM, CLUSTER, and ELEMENT and by FHIR whose extension mechanism allows for the definition of new attributes and structures within resource profiles. The motivation for this decision is, unlike FHIR and the OpenEHR models (both physical models), CIMI is a logical model and does not rely on extensions to provide for additional expressivity beyond what is provided by the CIMI Reference Model. The CIMI Logical Model favors expressivity over economy of structure and delegates model extensibility to its underlying physical model target. For instance, attributes existing in the CIMI Reference Model, but not existing in the FHIR Core Model, shall be mapped to the appropriate FHIR extensions.
The CIMI Reference Model is a layered model designed to be modular and currently consists of three layers:
This modular approach allows for additional, more domain-specific layers to be added in future; or for alternate iso-semantic patterns to be introduced at the appropriate level in the model. Over time, it can be expected the lower level reference model modules will become more stable while higher-level modules may still undergo additional flux.
A reference model pattern is a structural pattern (a single class or group of related classes) that can be constrained by archetypes in order to define a family of related and consistent detailed clinical models. The set of allowable clinical patterns comprise the CIMI Reference Model.
The CIMI archetype hierarchies form the second part of the CIMI model. These hierarchies serve two primary purposes:
Archetypes can specialize more general archetypes in ADL. They do so by progressively constraining the underlying reference model pattern in a manner consistent with and not contradictory to the constraints specified in archetypes higher up in the hierarchy.
Examples of constraint refinements are listed below:
Detailed Clinical Models (DCMs), highly specific models enabling the interoperable exchange of clinical information, typically reside at the leaf-level of CIMI archetype hierarchies. The cumulative constraints applied on a DCM are intended to be precise enough to allow for the unambiguous exchange of interoperable clinical information and thus constitute highly specific constraints on the underlying reference model pattern. This layered approach is illustrated below:
Figure 4: CIMI Architectural Framework
The CIMI reference model defines the core structure and data types of all CIMI clinical models. Each reference model module is further described in the sections below.
The CIMI Core Reference model module introduces four core reference model hierarchies:
Table XX below aligns the set of CIMI primitive types and their mappings to UML and FHIR. In CIMI all primitive types derive from the Any abstract class:
CIMI Primitives | UML Primitives | XML Primitives | FHIR Primitives |
Boolean | Boolean | -- | boolean |
Real | Real | -- | decimal |
Byte | -- | byte | Byte |
Integer | Integer | -- | integer |
Character | Character | -- | -- |
String | String | -- | string |
URI | -- | anyUri | uri |
List<T> | -- | -- | -- |
Array<T> | -- | -- | -- |
Any (root type) | -- | -- | Element |
Figure 5: CIMI Primitives
Figure 6 - CIMI Primitive Types
Table XXX below aligns the CIMI ‘complex’ data types and their mappings to FHIR. In CIMI, all complex data types derive from the DATA_VALUE abstract class:
CIMI Data Types | FHIR Primitives | FHIR Complex Types |
DATE | date | -- |
TIME | time | -- |
DATE_TIME | dateTime | -- |
INSTANT | instant | |
COUNT | -- | Count |
UNSIGNED_INTEGER_COUNT | UnsignedInt | |
POSITIVE_INTEGER_COUNT | PositiveInt | |
PROPORTION | -- | Ratio<Quantity> |
QUANTITY | -- | Quantity |
DURATION | -- | Duration |
INTERVAL_VALUE<T> | -- | Period, Range |
PLAIN_TEXT | string* | -- |
CODED_TEXT | -- | Coding |
URI_VALUE | uri | -- |
EHR_URI | uri | -- |
IDENTIFIER | -- | Identifier (partial match) |
YESNO | boolean* | -- |
PARSABLE | Base64Binary, Markdown | -- |
MULTIMEDIA | -- | Attachment (partial match) |
ORDINAL | -- | -- |
DATA_VALUE (root type) | -- | Element |
Figure 7 -Extended FHIR Primitive Types
CIMI and FHIR differ in the boundary between primitives and complex data types as shown in the table above. Moreover, a number of FHIR types do not have equivalents in CIMI. These include SampledData, though such types can be accommodated within the CIMI CLUSTER hierarchy.
Figure 8 - CIMI Data Value Types
All classes in CIMI, apart from CIMI primitives and complex types, derive from either the ASSOCIATION_CLASS or LOCATABLE classes. An ASSOCIATION_CLASS represents a qualified relationship between two LOCATABLEs. The source LOCATABLE is the class containing the association. The target LOCATABLE is generally specified by a target attribute of the association class as introduced in specializations of this class. For instance, Interpretation is a specialization of the ASSOCIATION_CLASS pattern. The source of the association, e.g. an Assertion class with an interpretation attribute, owns the association class. Interpretation then specifies one or more clinical statements as its target.
Figure 9 - CIMI Core Model
Figure 10 - Example specializations of ASSOCIATION CLASS
The following sections introduce some of the reference model patterns found in CIMI that build upon the CIMI Core Reference Model Module.
The CIMI Foundational Reference Model is composed of the following top-level hierarchies; CLUSTER, BASE_ENTITY and VIRTUAL CLUSTER, COMPOSITION, CONTENT, PARTY, PARTICIPATION, and PARTY_RELATIONSHIP.
Figure 11 - CIMI Foundation Package
Figure 12 - CIMI Party Package
Each hierarchy is described below.
The CLUSTER abstract class is the starting point for CIMI structures such as addresses, contact information, and person names. The ENTITY specialization of CLUSTER is the parent classes for entities such as organizations, persons, medications, and devices. Unlike CLUSTER, VIRTUAL_CLUSTER allows for the grouping of attributes to support model component reuse and consistency but whose containment structure may be ignored by tools, editors, and code generation frameworks.
The CLUSTER hierarchy differs from the DATA_VALUE hierarchy in that specializations of DATA_VALUE represent a concise set of data types (i.e., entirely defined by the values they take; e.g., a code, a quantity or a proportion) while specializations of CLUSTER are used to define the far more numerous and variable instance structures used to compose the reference model patterns (e.g., person names or international address structures).
At this time, the COMPOSITION hierarchy consists of a single class that can be used to represent clinical reports or patient records at the archetype level. A composition is composed of sections and content entries.
The CONTENT hierarchy plays a special role in CIMI because it is the parent hierarchy for Clinical Statements. The CONTENT class has the following subclasses: SECTION and ENTRY. ENTRY, in turn, has two specializations: COMPOUND_ENTRY and INDIVISIBLE_ENTRY. A COMPOUND_ENTRY can be composed of other COMPOUND_ENTRYs and INDIVISIBLE_ENTRYs thus supporting a recursive pattern that can be constrained accordingly at the archetype level. ENTRYs represent units of standalone clinical information. An example of a COMPOUND_ENTRY may be a laboratory panel or a time series while an individual analyte or simple procedure may be represented by an INDIVISIBLE_ENTRY. Complex medication orders that are ordered as a unit (e.g., an insulin sliding scale or an oral steroid taper) are represented as INDIVISIBLE_ENTRYs with a potentially composite internal structure.
The SECTION content type may be used to represent sections in a document or a simple collection of entries without metadata. Note the latter use of SECTION differs from COMPOUND_ENTRY representing a logical grouping of entries that can hold metadata about the grouping. For instance, a laboratory panel is a type of COMPOUND_ENTRY holding provenance information for the panel as a whole.
The CIMI Foundational Reference Model introduces the participation pattern. It defines PARTY, which may be either a ROLE or an ACTOR, and a PARTY_RELATIONSHIP to represent a relationship between two parties such as an ACTOR playing one or more roles in the performance of an activity. Each class in the pattern has a type attribute to allow the binding of formal semantics at the archetype level – concepts representing the type of actor or the type of role performed. These classes also serve as the root types for specializations introduced in other reference modules. It is important to note ACTORs are typically ENTITYs acting within a well-defined ROLE in a PARTICIPATION. For instance, a procedure may be performed by a John Smith (ACTOR) acting as a clinician (ROLE) in a surgeon PARTICIPATION.
The CIMI Clinical Reference Model module includes the classes and structural patterns derived from those classes upon which all CIMI archetypes are built.
The CIMI Clinical Reference Model Party pattern builds upon the CIMI Foundational Reference Model PARTY pattern as shown below. It adds a number of attributes to ROLE and defines two role specializations: a HealthCareConsumerRole and a HealthCareProviderRole. The association between a person or organization and the role they play in an activity is achieved by the use of PARTY_RELATIONSHIP and PARTICIPATION classes (or specializations thereof) defined in the CIMI Foundational Reference Model module.
Figure 13 - Specializations of the PARTY class
The CIMI Clinical Statement Pattern forms the core of the CIMI model. The BaseClinicalStatement class is a specialization of the ENTRY class from the Foundation Reference Model module. It represents a statement about some aspect of a health care process. The CIMI ClinicalStatement pattern contains a statement topic (StatementTopic) and the situational context for that topic (StatementContext). The BaseClinicalStatement pattern also includes relevant attribution metadata for the information contained in a clinical statement (please refer to the Attribution/Provenance Patterns section below). Clinical Statements may either by Indivisible Clinical Statements or Composite Clinical Statements (e.g., a panel).
Figure 14 - Clinical Statement Pattern
The StatementTopic abstract class has three specializations:
Note for the May 2017 ballot cycle we limit the scope of the model to the statement topics presented below. More topic specializations will be added over time.
Figure 15 – Clinical Statement Topics
The StatementContext abstract class has the following three specializations:
Consider a Proposal context having the following lifecycle states: initiated, reviewed, updated, approved, and rejected. Attribution information for each of these activities is captured by the ClinicalStatement.contextStatusHistory attribute while the latest status of the proposal action context is captured by the attribute ActionContext.currentStatus of type CODED_TEXT. The value of the currentStatus attribute is the Attribution.activity code associated the last status attribution in the context status history for the given context. The ClinicalStatement.contextAttribution provides attribution information for the context itself (e.g., who initiated the proposal, when, how, etc…) and typically corresponds to the first status in the context status history. This is illustrated in the diagram below. Each line represents an activity moving the proposal along one of its lifecycle states. The graph itself represents the set of allowed paths. The blue lines represent the path traversed in the lifecycle of this proposal. Each dotted line represents a valid transition though not one followed by this proposal.
Figure 16 – Proposal Lifecycle
The ClinicalStatement.contextStatusHistory is the ordered list of attributions associated with each transition moving from start to finish. It consists of the following list:
State Transition attributions | Value of Attribution.activity attribute |
Initiate (also the context attribution) | “initiate” |
Review | “review” |
Approve | “approve” (also the context’s current status) |
Figure 17 – State Transition Attribute
The ClinicalStatement.contextAttribution refers to the Initiate Attribution in this context status history list.
The ActionContext.currentStatus refers to the activity code of the last transition followed by the proposal. In this case, it is the activity code of the Approve attribution or the “approve” code in this illustration.
Figure 18 - ClinicalStatement Contexts
Figure 19 - Finding Contexts
Figure 20 - Action Contexts
StatementTopic and StatementContext are attribute groups (VIRTUAL_CLUSTERs) and have the following characteristics:
The CIMI ClinicalStatement pattern aligns with the SNOMED CT Situation with Explicit Context Concept Model as defined in the SNOMED CT Editorial Guide[6]. The attributes are as follows:
In the CIMI model, provenance information is represented by the Attribution class. The Attribution class provides a pattern for the capture of provenance information such as the what, who, when, where, why, and how associated with a particular activity – e.g., provenance attributes about the verification of a clinical statement.
CIMI currently includes two attribution patterns:
Figure 21 - Provenance Patterns
At this time, CIMI defines two specialization of the Finding Statement Topic: Assertion and EvaluationResult.
The assertion model is used to capture information about a unary clinical finding, whereas the evaluation result model is used to capture a value ascertained about an observable. An evaluation result consists of an observed characteristic (sometimes referred to as ‘the question’) and a value for the observed characteristic (sometimes referred to as ‘the answer’). At this time, a number of specializations for Assertion have been proposed (BradenAssessment, FindingSiteAssertion, WoundAssertion, DeviceAssertion, and RiskAssertion) and we anticipate additional specializations in the future. Two specializations of Evaluation Result are provided in the reference model: LaboratoryTestResult and PhysicalEvaluationResult. From these, a number of core archetypes are provided such as QuantitativeLaboratoryTestResult and CodedLaboratoryTestResult. These archetypes constrain the result attribute of EvaluationResult to a QUANTITY and a CODED_TEXT respectively. Please refer to the archetypes for more examples of such constraints.
For more information on the proper usage of these classes, please refer to the Style Guide section below.
Both models are shown below.
Figure 22 – The Assertion Statement Topic
Figure 23 – The EvaluationResult Statement Topics
Figure 24 – Wound Assertion
As part of this submission we include an example of the Act statement topic, namely Procedure and some proposed specializations including LaboratoryProcedure, SpecimenCollection, and SurgicalProcedure. Note these models are still incomplete at this time but serve as examples of clinical statements about actions and the expression of ‘negation’ patterns in CIMI (e.g., ProcedureNotPerformed is composed of the Procedure statement topic and the NonPerformance statement context).
Figure 25 - Action Topics
Cimi entities represent material entities such as substances, medications, devices, or people as well as non-material entities such as organizations. Entities may be actors in participations. Entities and their related structures are shown below:
Figure 26 – The Entity Hierarchy
Common CIMI Data Structures are reusable components derived from the CLUSTER supertype which are shared by a number of other classes. They are used to define component structures necessary in the construction of CIMI patterns. Examples include structures such as Address, PartyName, BirthData, and so on. For more information on these structures, please refer to the diagrams below and to the CIMI Logical Model Specification.
Figure 28 - Address Structures
Figure 29 - Party and Related Structures
The CIMI Clinical Reference Model module defines a number of association classes based on the ASSOCIATION_CLASS pattern described in the CIMI Core Reference Model module section of this document. Association classes allow the qualification of relationships and are useful when a relationship between a source and one or more targets specifies additional attributes. Examples of such association classes are the Justification and Interpretation classes, both of which derive from the ClinicalStatementAssociation class. The Association Class pattern is illustrated below:
Figure 30 - Party and Related Structures
While the ASSOCIATION_CLASS in the Core Reference Model does not define a target attribute, it is expected specializations of the class provide a target attribute for the subtype of locatable most appropriate for the use case.
Cimi archetypes include complete “detailed clinical models” fit for use as well as their ancestors in a hierarchy of constraints. At the top of this hierarchy are CIMI top-level archetypes from which all CIMI archetypes are derived.
CIMI Top-Level archetypes define the following functions:
Each archetype in a hierarchy is by definition consistent with all its ancestors.
In this section we introduce the archetype hierarchies and provide example archetypes for each of the functions described above.
For this submission, the following archetype hierarchies have been developed.
The Attribution hierarchy includes a number of specializations of the Attribution reference model pattern useful for archetype development and for capturing provenance information about activities. These include archetypes for the following activities: author, record, sign, and verify referenced in clinical statement archetypes. These archetypes illustrate the use_archetype pattern, whereby an archetype can reference another archetype in its definition. Please note, however, Attribution archetypes do not yet specify the required terminology bindings for coded attributes. These bindings will be addressed in a later submission.
Figure 31 - Attribution Archetype Hierarchy
The example below shows how the base_clinical_statement archetype refines the reference model BaseClinicalStatement class by constraining the ClinicalStatement.authored attribute of type Attribution to those constraints specified by the author_action archetype:
archetype (adl_version=2.0.6; rm_release=2.0.2; generated)
CIMI-CORE-BaseClinicalStatement.base_clinical_statement.v1.0.0
definition
BaseClinicalStatement[id1] matches {
...
authored matches {
use_archetype Attribution [id8, CIMI-CORE-Attribution.author_action.v1]
}
recorded matches {
use_archetype Attribution [id9, CIMI-CORE-Attribution.record_action.v1]
}
verified matches {
use_archetype Attribution [id10, CIMI-CORE-Attribution.verify_action.v1]
}
signed matches {
use_archetype Attribution [id11, CIMI-CORE-Attribution.sign_action.v1]
}
cosigned matches {
use_archetype Attribution [id12, CIMI-CORE-Attribution.sign_action.v1]
}
...
}
The StatementContext hierarchy specializes CIMI clinical statement contexts in conformance with the SNOMED CT Situation with Explicit Context. This hierarchy has two branches:
Additional specializations of the statement_context archetype will be added over time.
Figure 32 - StatementContext Hierarchy
Statement context archetypes promote a compositional (and reusable) approach for specifying the context of a clinical statement’s topic such as whether a finding was present or absent or an action performed or not performed (see Clinical Statement Pattern below). They also provide important attribution information which can be appropriately constrained in specializations. The statement_context archetype initially bind the meaning of StatementContext attributes to the corresponding SNOMED CT Situation With Explicit Context concept model attributes. It also defines high level range constraints for the context attribute. The specializations of the statement_context archetype progressively constrain the set of allowable values for the context attribute. For instance, the ActionContext specifies that the context attribute’s range must be a descendant of the concept | Context values for actions | 288532009 (<=)(< Q). The Proposal archetype further constrains this range to a single code |Under consideration (qualifier value)| 385642001 (==).
The Statement Topic hierarchy will probably be the most active hierarchy in CIMI. The StatementTopic class and archetype specializations represents the core clinical expressivity of a domain. At this time only a few specializations have been introduced. More will be added over time. They are listed below:
Figure 33 - StatementTopic Hierarchy
Archetype constraints on the BaseClinicalStatement reference model pattern form the core of the CIMI model. As part of this submission we include a number of illustrative though partially complete clinical statement archetypes. They fall into two main categories:
The indivisible clinical statement hierarchy is illustrated below:
Figure 34 - Clinical Statement Hierarchy
Examples of Compound Clinical Statements are shown below:
Figure 35 - Compound Clinical Statement
The action clinical statement archetype hierarchy is developed by progressively constraining the context of the Clinical_Statement core archetypes along the indivisible and compound clinical statement axes as shown below:
Figure 36 - Indivisible Clinical Statement
A similar approach is taken for clinical statements about findings:
Figure 37 - Finding Statements
At this time the StatementTopic and StatementContext hierarchies are the only two CLUSTER archetype hierarchies defined. In the future archetypes will be developed for all reference model classes originating from CLUSTER.
In the following sections we elaborate on the functions of archetype hierarchies.
Top level archetypes often constrain the range of coded attributes by binding them to intentionally defined value sets. For instance, the statement_context archetype constrains its context attribute to the intentional value set resulting from the expression < 288529006 |Context values (qualifier value)|. Any specialization of this archetype must define value set constraints on the context attribute consistent with this intentional definition. That is, all downstream value set constraints must be subsets of the value set expansion that result from the evaluation of the above expression. The action_context archetype specifies the following constraint on the context attribute: < 288532009 |Context values for actions (qualifier value)|. The specified constraint is valid because the concept 288532009 is properly subsumed by the concept 288529006. In turn, the specialization proposal of action_context further constrains the attribute context to the following expression: = 385642001 |Under consideration (qualifier value)| (i.e., a value set consisting of a single code). This too is a valid constraint since this one-code value set is subsumed by all parent value sets defined above it.
This is illustrated below:
Figure 38 - Progressive applications of Attribute Range Constraints
The CIMI EvaluationResult Statement Topic pattern specifies the result of an evaluation has as its type the abstract class DATA_VALUE. Doing so enables modelers to constrain the type of the EvaluationResult.result attribute into any valid specialization of DATA_VALUE. Such an archetype pattern allows modelers to define families of models entirely within the archetype layer of the CIMI model such as a quantitative_physical_evaluation_result which constrains the result attribute to a QUANTITY type (a valid subtype of DATA_VALUE). Similarly, modelers may wish to create a coded_physical_evaluation_result which constrains the result attribute of EvaluationResult (or any of its reference model specializations) to a CODED_TEXT. Further archetype specializations can be defined. Note a subtype cannot relax a parent constraint. For instance, a specialization of coded_physical_evaluation_result cannot set the type of the result attribute as TEXT since TEXT is the supertype of CODED_TEXT and such a constraint would loosen rather than tighten the parent constraint. This is illustrated below:
Figure 39 - Progressive applications of Attribute Type Constraints
As described earlier in this document, the BaseClinicalStatement reference model pattern is one of the core patterns of the CIMI model. However, the BaseClinicalStatement (including its two specializations IndivisibleClinicalStatement and CompoundClinicalStatement) pattern only becomes meaningful through the archetype patterns derived from it.
The StatementTopic, StatementContext, and BaseClinicalStatement classes serve as the reference model patterns for three top level archetype hierarchies:
This approach is illustrated below:
Figure 40 - The Clinical Statement-Statement Topic-Statement Context Pattern
The act_statement archetype definition is shown below:
IndivisibleClinicalStatement[id1.1.1] matches {
topic matches {
use_archetype Act [id4.0.1,
CIMI-CORE-Act.act.v1]
}
context matches {
use_archetype ActionContext [id5.0.1,
CIMI-CORE-ActionContext.action_context.v1]
}
}
[1] http://www.openehr.org/releases/AM/latest/docs/AOM2/AOM2.html
[4] http://www.openehr.org/releases/BASE/latest/docs/bmm/bmm.html
[5] http://www.openehr.org/releases/AM/latest/docs/ADL2/ADL2.html
[6] https://confluence.ihtsdotools.org/display/DOCEG